|
Plenary
Lecture
Resolving Nanoscale Details of Ligands at their Binding
Sites of Membrane Targets

Professor Anthony Watts
Biomembrane Structure Unit
Biochemistry Department
Oxford University, Oxford
OX1 3QU, UK
E-mail:
anthony.watts@bioch.ox.ac.uk
Abstract: It is now
possible to resolve local dynamics
within a membrane bound protein at
near physiological conditions in
natural membrane fragments or in
reconstituted complexes, using
solid state NMR approaches [1, 2].
This information is obtained by
isotopically (2H, 13C, 19F, 15N,
17O) labeling selective parts of
either a ligand or the protein
understudy, and observing the
nucleus in non-crystalline,
macromolecular complexes [3,4].
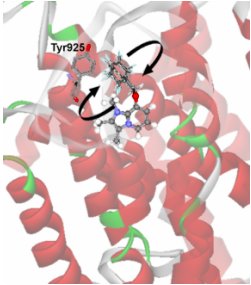
Ligands with complex structure
have differential mobility at
their binding sites. Substituted
imidazole pyridines, for example,
which inhibit the H+/K+-ATPase and
have therapeutic use, are
constrained in the imidazole
moiety, but shows significant
flexibility at the pyridine group
[5] (see figure). It is this group
which has a direct interaction
with an aromatic (phe198) residue,
with the potential for ?-electron
sharing [6]. Similarly, the
steroid moiety of ouabain
undergoes motions which are
similar to those of the target
protein, the Na+/K+-ATPase, but
the rhamnose undergoes a high
degree of flexibility at fast
rates of motions whilst
interacting with Tyr198 [7]. For
acetyl choline when bound in the
nicotinic acetyl choline receptor
(nAChR), the quaternary ammonium
group undergoes fast rotation at
an aromatic binding site, which is
driven by thermal fluctuations
which may be functionally
significant [8]. Our current focus
is on GPCRs, specifically the
brain neurotensin receptor (NTS1)
for which the structure (by single
molecule cryo-EM) and ligand
binding interactions are being
studied [9 - 14].
|

